Lecture 3 Basic Visualization
This lecture goes over basic visualization methods. Objectives are the following:
-Visualize Slices
-Digital Zoom
-Histograms of Intensities
-Back mapping
3.1 Read NIfTI
## oro.nifti 0.10.1dir = "Neurohacking_data/BRAINIX/NIfTI/"
fname = "Output_3D_File"
fpath = paste0(dir,fname)
(nii_T1 = readNIfTI(fname = fpath))## NIfTI-1 format
## Type : nifti
## Data Type : 4 (INT16)
## Bits per Pixel : 16
## Slice Code : 0 (Unknown)
## Intent Code : 0 (None)
## Qform Code : 2 (Aligned_Anat)
## Sform Code : 2 (Aligned_Anat)
## Dimension : 512 x 512 x 22
## Pixel Dimension : 0.47 x 0.47 x 5
## Voxel Units : mm
## Time Units : secPrinting the NIfTI object show the structure of the data
3.2 Visualizing Slices
As a first pass, we could use the image function from the graphics package.
#Save dimensions of the image
d = dim(nii_T1)
# Visualizing the 11th axial slice
graphics::image(1:d[1], 1:d[2], nii_T1[,,11],
col = gray(0:64/64), xlab="", ylab="") 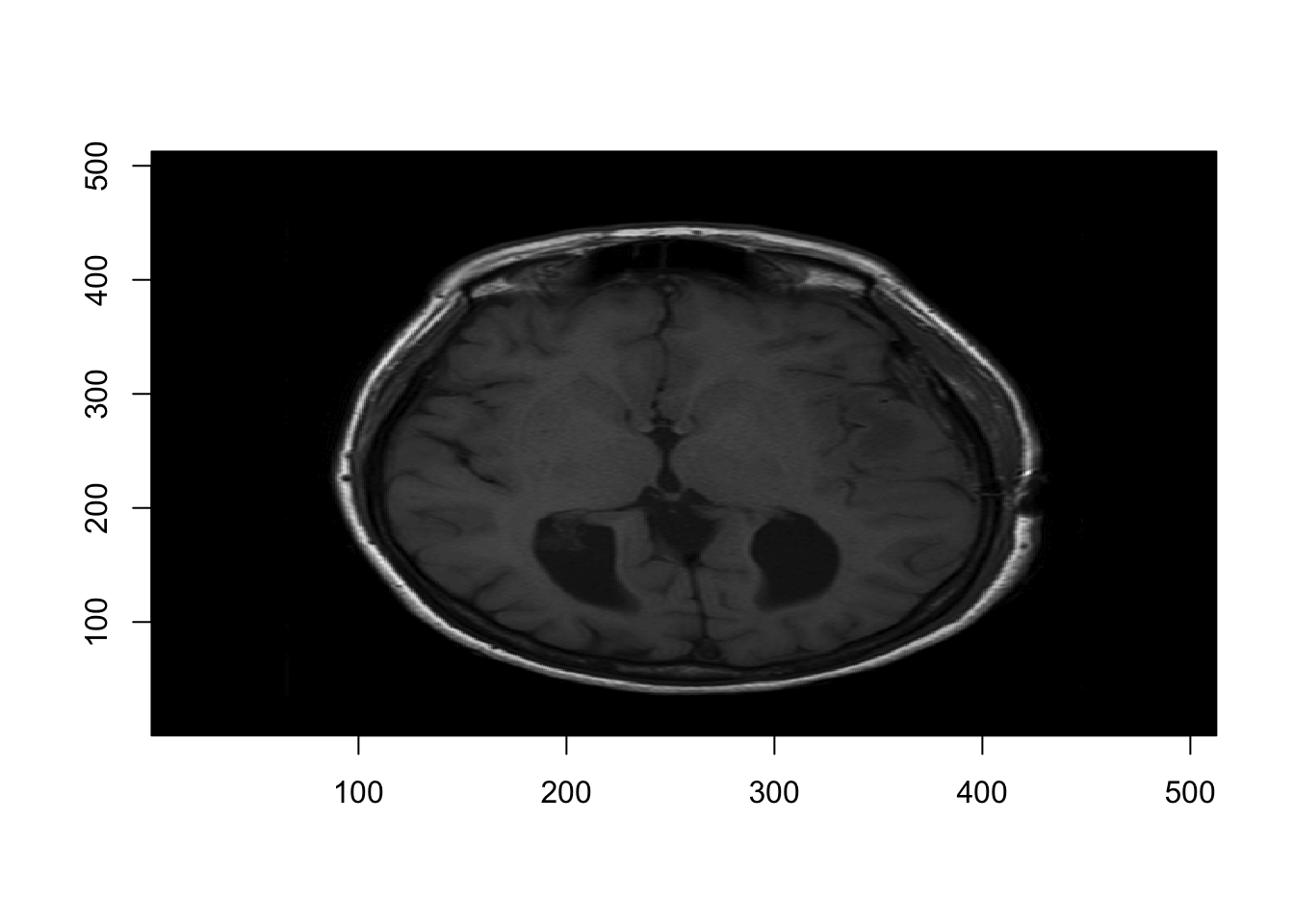
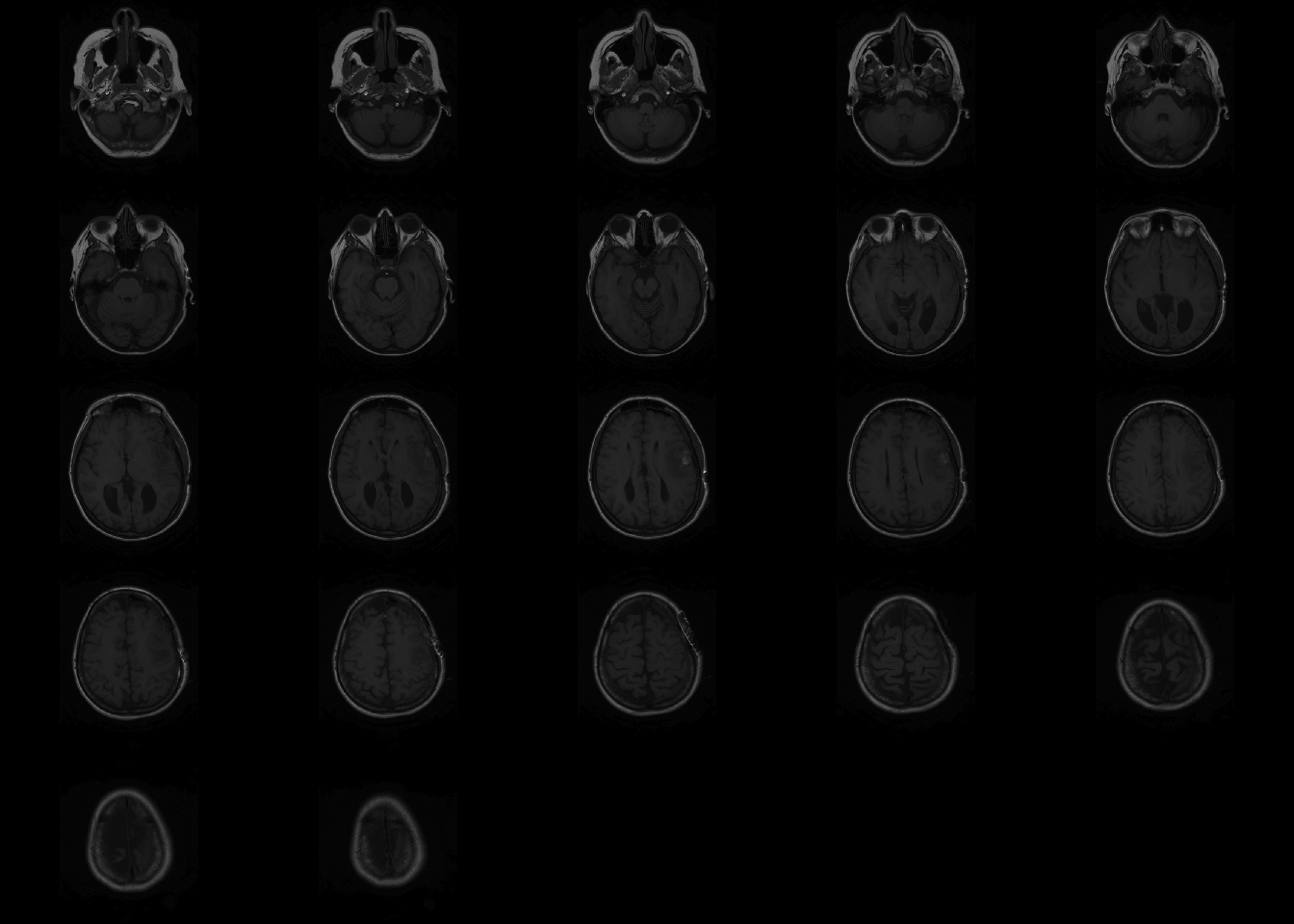
However, oro.nifti has its own image function which is called by default one NIfTI objects.
If no arguments are passed for coordinates, oro.nifti::image plots all the slices axially.
The orthographic functions allows to visualize a single point from three dimensions.

3.3 Exploratory histogram of intensities
Using R base graphics, we can get a quick view of the intensites in the brain.
par(mfrow = c(1, 2));
o = par(mar = c(4,4,0,0))
hist(nii_T1, breaks = 75, prob = T, xlab="T1 intensities", col = rgb(0,0,1, 1/2), main="")
# Plot filtered by > 20
hist(nii_T1[nii_T1 > 20], breaks = 75, prob = T, xlab="T1 intensities > 20", col = rgb(0,0,1, 1/2), main="" )
A lot ot the voxels, are ~0 in the first plot since most of the image is the black background. The second plot shows the intensities that are higher than 20.
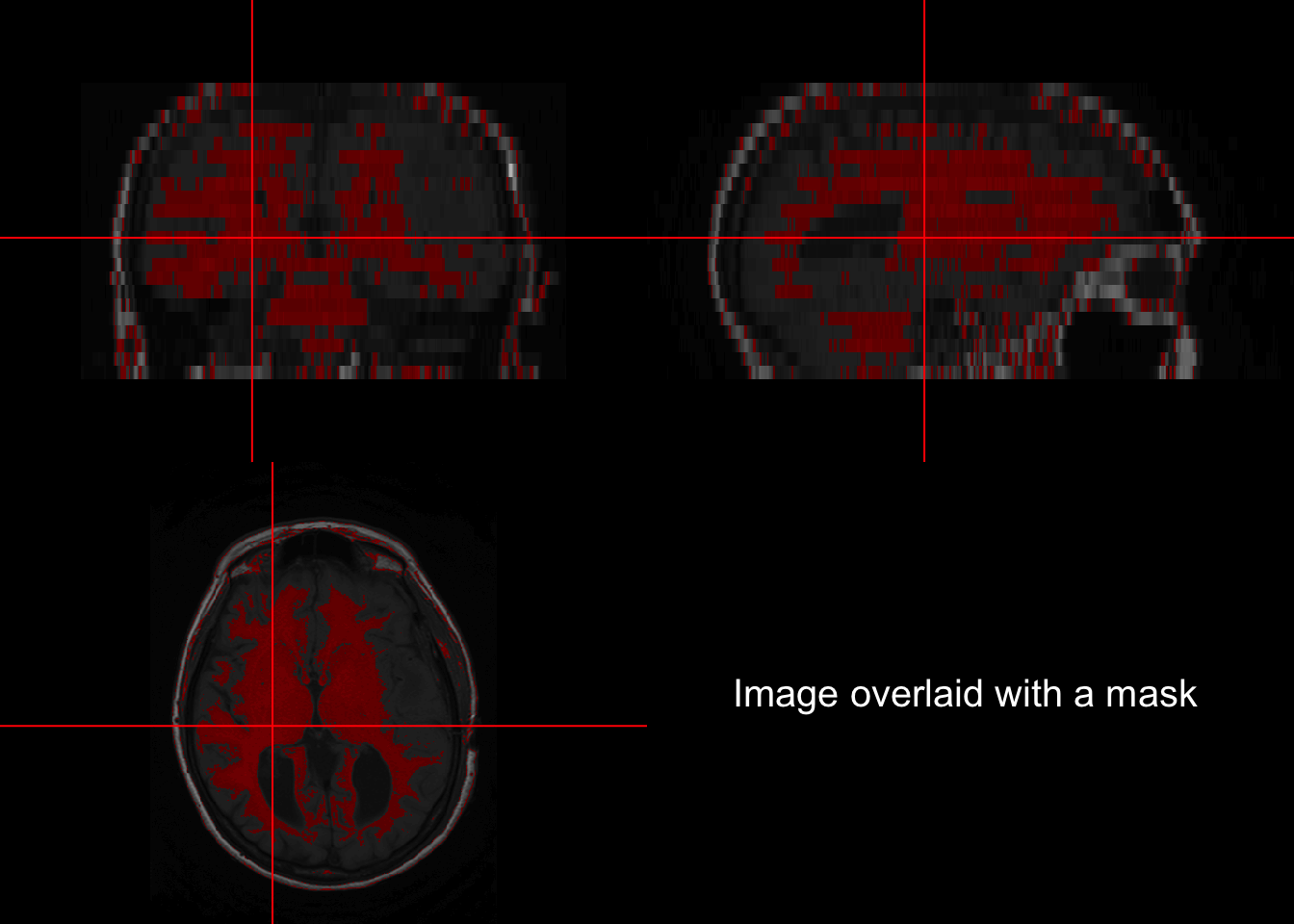
3.4 Back mapping
Backmapping consists of overlaying a mask over an image of the brain. A basic backmapping can be achieved by taking creating a mask according to intensities and calling the function overlay.
is_btw_300_400 = nii_T1 > 300 & nii_T1 < 400
nii_T1_mask = nii_T1
nii_T1_mask[!is_btw_300_400] = NA
overlay(nii_T1, nii_T1_mask, z = 11, plot.type="single")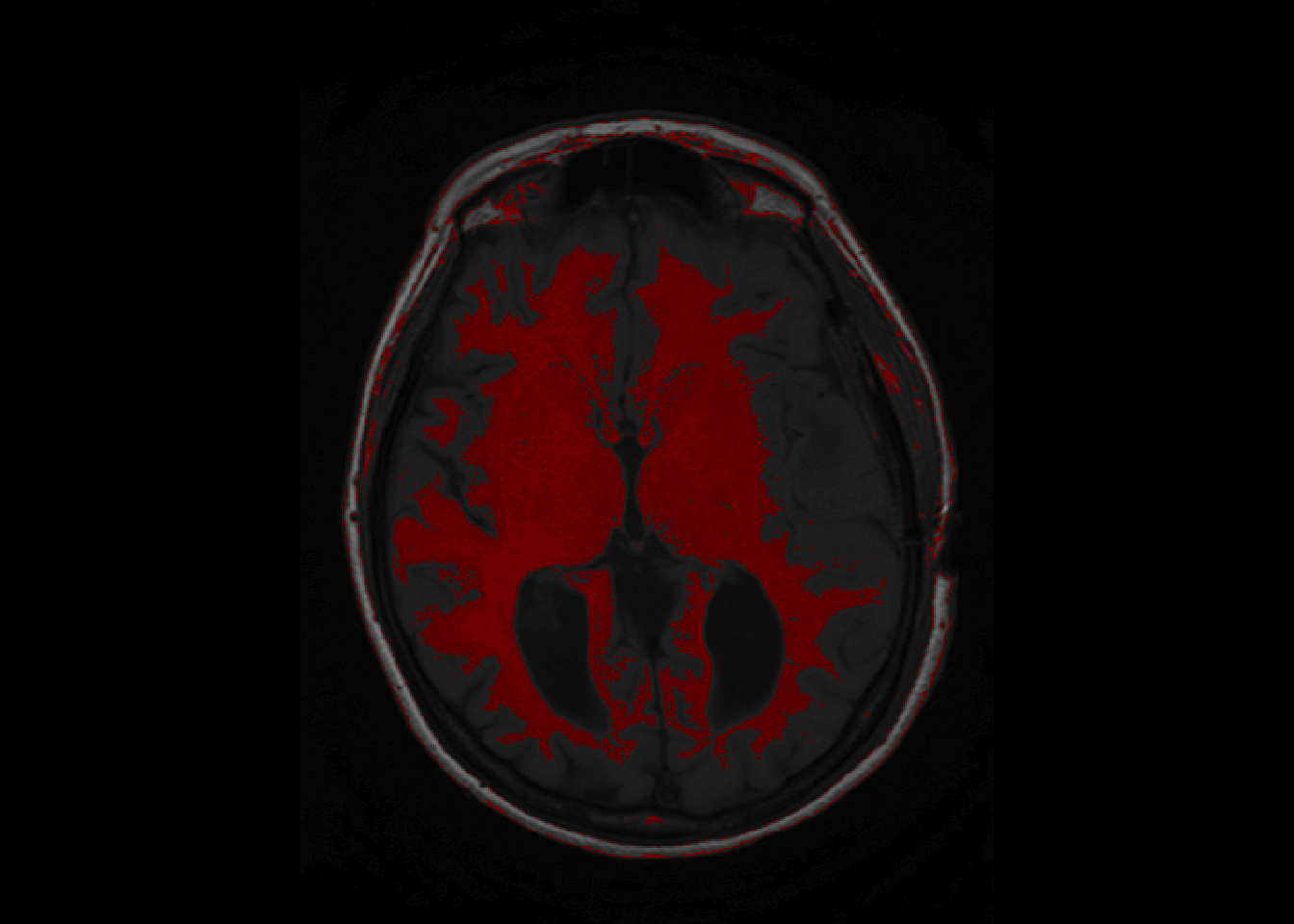
overlay can be called to show a single slice as above, or the whole brain as below.
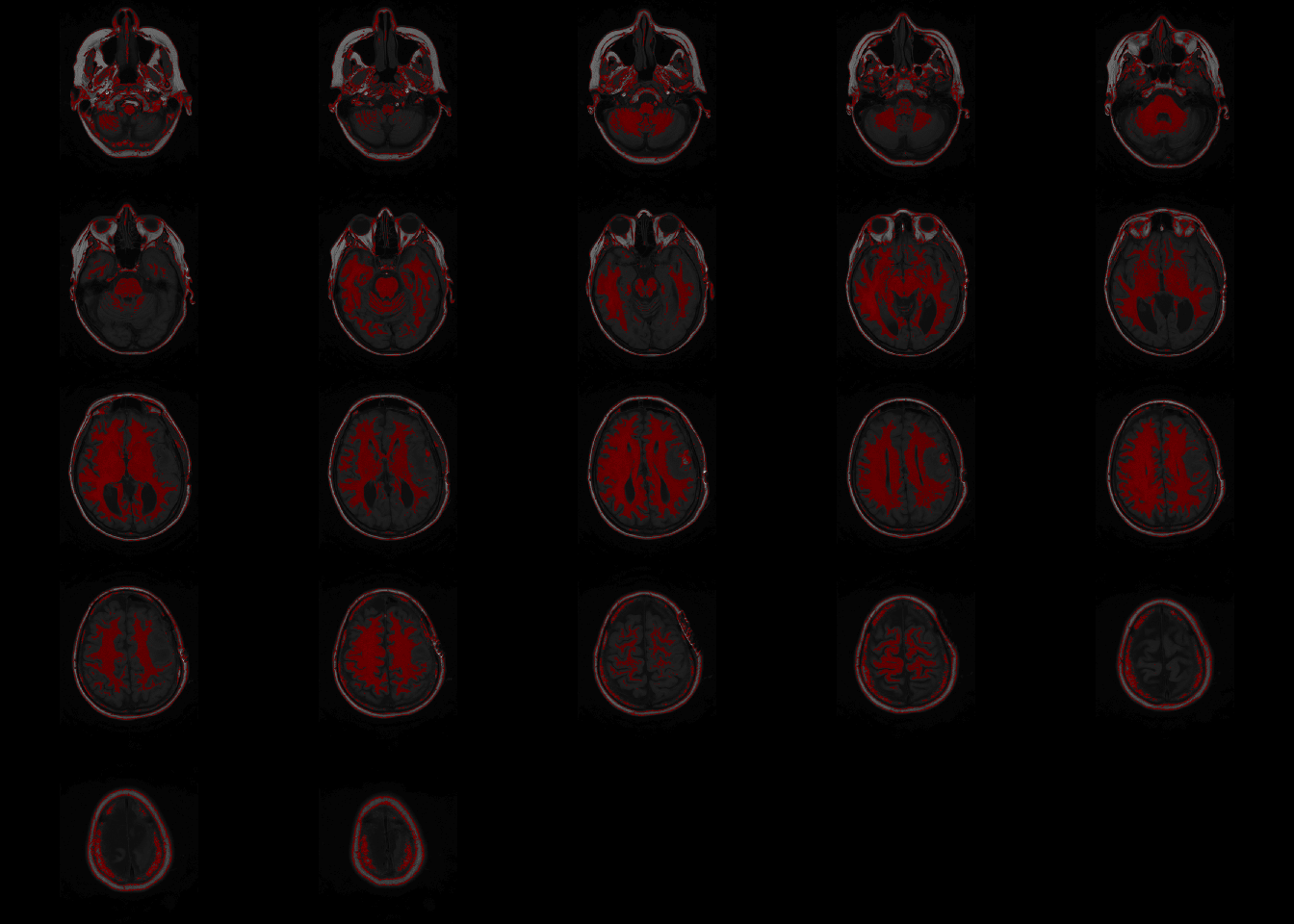
orthographic(nii_T1, nii_T1_mask, xyz=c(200,220,11), text="Image overlaid with a mask", text.cex = 1.5)## Warning in min(x, na.rm = na.rm): no non-missing arguments to min; returning Inf## Warning in max(x, na.rm = na.rm): no non-missing arguments to max; returning -
## Inf